Human tissue segmentation
| GitHub repository |
|---|
Summary
Kaggle's HuBMAP + HPA - Hacking the Human Body competition involves segmenting functional tissue units (FTUs) in images of human tissue microscope slides. The top 5 submissions to the project finetune a pretrained UNet model. I investigated whether at DeepLabV3 model might perform better at the task, since this newer model is designed to be particularly robust to changes in scale, which are common in microscopy. While I found DeepLabV3 to be unsuitable, I found that transferring the same hyperparameters used for my best DeepLabV3 model to a UNet architecture resulted in a model that ranked in the top 5 of Kaggle submissions on the very first attempt.
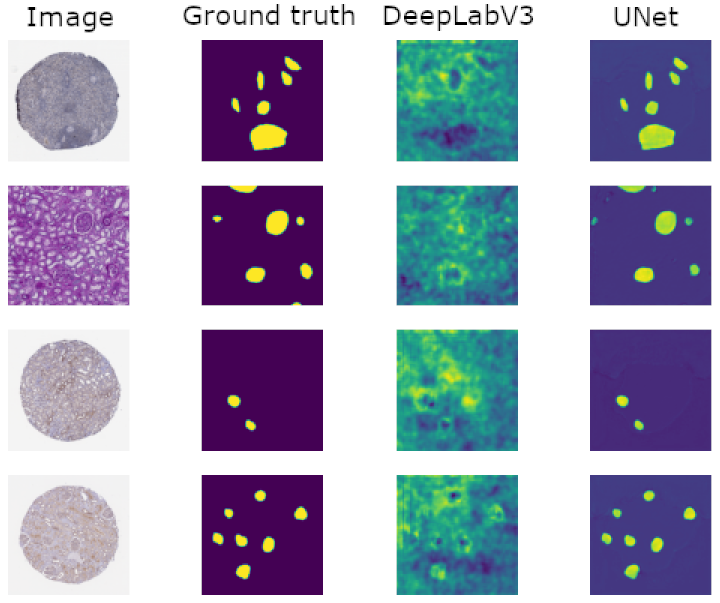
The figure below shows four slides from the validation dataset, along with the ground truth segmentation labels. As can be seen, my UNet model segments the samples to a high standard of accuracy, even though the corresponding DeepLabV3 model performed poorly.
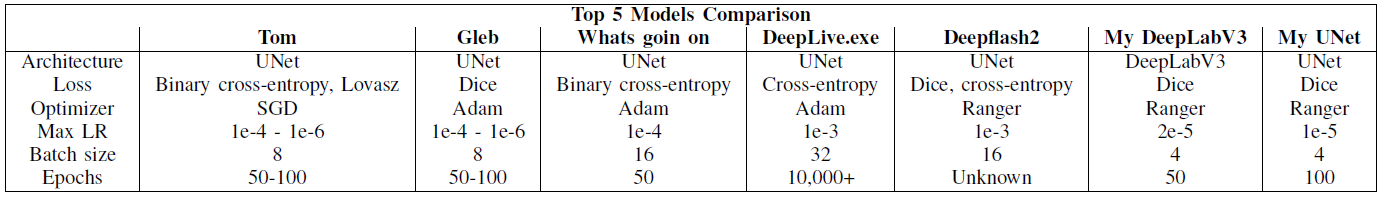
Below is a comparison of my models' hyperparameters (rightmost two columns) with the hyperparameters used in the top 5 solutions.
Skills gained
I learned how to work with the Ranger optimizer, which improves on the popular Adam optimizer by adding lookahead. Through consulting a PhD student in microbiology at Harvard, I also learned best practices for computer vision on stained microscopy slides. For example, pixel normalization, commonly used elsewhere in computer vision, should not be used in microscopy because the exact hues of the dyes convey useful information.